Bagging, Boosting, and Random Forests
[math] \require{boldsymbol} \DeclareMathOperator*{\argmax}{argmax} \newcommand{\D}{\mathcal{D}_n} \newcommand{\T}{\Theta} \newcommand{\Pt}{\mathbb{P}_{\Theta}} \newcommand{\Vt}{\mathbb{V}_{\Theta}} \newcommand{\Et}{\mathbb{E}_{\Theta}} \newcommand{\E}{\mathbb{E}} \newcommand{\V}{\mathbb{V}} \renewcommand{\P}{\mathbb{P}} \newcommand{\LL}{\mathbb{L}} \newcommand{\Ccal}{\mathcal{C}} \newcommand{\Pcal}{\mathcal{P}} \newcommand{\N}{\mathbb{N}} \newcommand{\e}{\varepsilon} \newcommand{\R}{\mathbb{R}} \newcommand{\bX}{{\bf X}} \newcommand{\bx}{{\bf x}} \newcommand{\bz}{{\bf z}} \newcommand{\smallO}[1]{\ensuremath{\mathop{}\mathopen{}o\mathopen{}\left(#1\right)}} \newcommand{\Mtry}{\mathcal{M}_{\textrm{try}}} \newcommand{\Pfin}{\mathcal{P}_{\textrm{final}}} \newcommand{\mtry}{\texttt{mtry}} \newcommand{\bm}[1]{\boldsymbol{#1}} [/math]
Bootstrap aggregating, also called bagging (from bootstrap aggregating), is a machine learning ensemble meta-algorithm designed to improve the stability and accuracy of machine learning algorithms used in statistical classification and regression. It also reduces variance and helps to avoid overfitting. Although it is usually applied to decision tree methods, it can be used with any type of method. Bagging is a special case of the model averaging approach.
Description of the technique
Given a standard training set [math]D[/math] of size [math]n[/math], bagging generates [math]m[/math] new training sets [math]D_i[/math], each of size [math]n[/math], by sampling from [math]D[/math] uniformly and with replacement. By sampling with replacement, some observations may be repeated in each [math]D_i[/math]. If [math]n=n'[/math], then for large [math]n[/math] the set [math]D_i[/math] is expected to have the fraction (1 - 1/e) (≈63.2%) of the unique examples of [math]D[/math], the rest being duplicates.[1] This kind of sample is known as a bootstrap sample. Sampling with replacement ensures each bootstrap is independent from its peers, as it does not depend on previous chosen samples when sampling. Then, [math]m[/math] models are fitted using the above [math]m[/math] bootstrap samples and combined by averaging the output (for regression) or voting (for classification).

Bagging leads to "improvements for unstable procedures",[2] which include, for example, artificial neural networks, classification and regression trees, and subset selection in linear regression.[3] Bagging was shown to improve preimage learning.[4][5] On the other hand, it can mildly degrade the performance of stable methods such as K-nearest neighbors.[2]
Creating the bootstrap dataset
The bootstrap dataset is made by randomly picking objects from the original dataset. Also, it must be the same size as the original dataset. However, the difference is that the bootstrap dataset can have duplicate objects. Here is simple example to demonstrate how it works along with the illustration below:
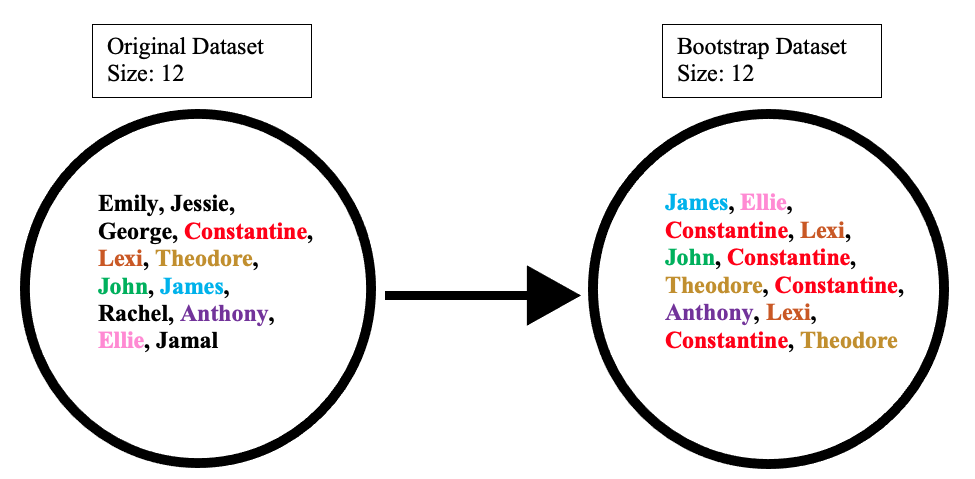
Suppose the original dataset is a group of 12 people. These guys are Emily, Jessie, George, Constantine, Lexi, Theodore, John, James, Rachel, Anthony, Ellie, and Jamal.
By randomly picking a group of names, let us say our bootstrap dataset had James, Ellie, Constantine, Lexi, John, Constantine, Theodore, Constantine, Anthony, Lexi, Constantine, and Theodore. In this case, the bootstrap sample contained four duplicates for Constantine, and two duplicates for Lexi, and Theodore.
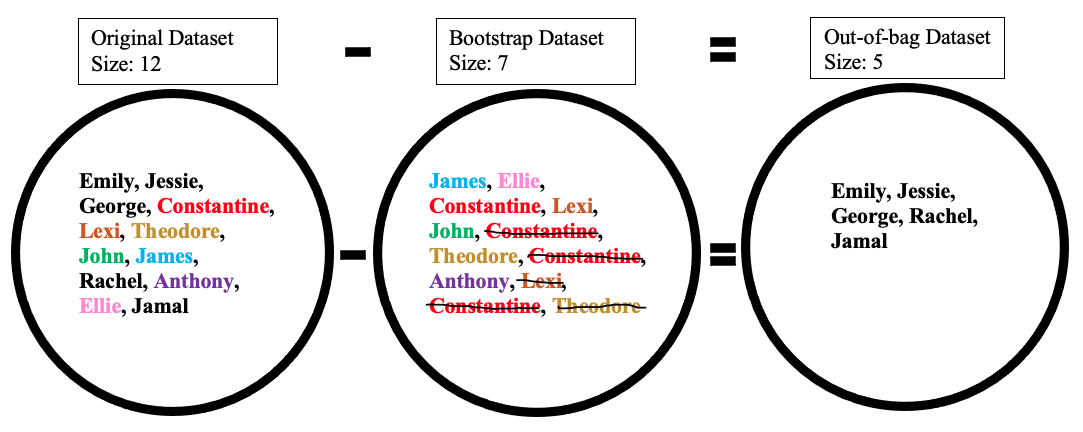
Creating the out-of-bag dataset
The out-of-bag dataset represents the remaining people who were not in the bootstrap dataset. It can be calculated by taking the difference between the original and the bootstrap datasets. In this case, the remaining samples who were not selected are Emily, Jessie, George, Rachel, and Jamal. Keep in mind that since both datasets are sets, when taking the difference the duplicate names are ignored in the bootstrap dataset. The illustration below shows how the math is done:
Example: ozone data
To illustrate the basic principles of bagging, below is an analysis on the relationship between ozone and temperature (data from Rousseeuw and Leroy (1986), analysis done in R).
The relationship between temperature and ozone appears to be nonlinear in this data set, based on the scatter plot. To mathematically describe this relationship, LOESS smoothers (with bandwidth 0.5) are used. Rather than building a single smoother for the complete data set, 100 bootstrap samples were drawn. Each sample is composed of a random subset of the original data and maintains a semblance of the master set’s distribution and variability. For each bootstrap sample, a LOESS smoother was fit. Predictions from these 100 smoothers were then made across the range of the data. The black lines represent these initial predictions. The lines lack agreement in their predictions and tend to overfit their data points: evident by the wobbly flow of the lines.

By taking the average of 100 smoothers, each corresponding to a subset of the original data set, we arrive at one bagged predictor (red line). The red line's flow is stable and does not overly conform to any data point(s).
Boosting
This section is based on [6]. The original boosting algorithm [7] emerged from the field of machine learning, where it gained much interest and was soon considered as a powerful instrument to predict binary outcomes. The basic idea is to iteratively apply simple classifiers and to combine their solutions to obtain a better prediction result. The concept of boosting was later adapted to the field of statistical modelling, where it can be used to select and estimate the effect of predictors on a univariate response variable in different types of regression settings [8].
The reasons for the success of statistical boosting algorithms are
- their ability to incorporate automated variable selection and model choice in the fitting process,
- their flexibility regarding the type of predictor effects that can be included in the final model and
- their stability in the case of high-dimensional data with possibly far more candidate variables than observations -- a setting where most conventional estimation algorithms for regression settings collapse.
The concept of boosting emerged from the field of supervised learning, which is the automated learning of an algorithm based on labelled data with observed outcome in order to make valid predictions for unlabelled future or unobserved data. Supervised learning is a subdiscipline of {\em machine learning}, which also comprises unsupervised learning based on unlabelled data and semi-supervised learning which is a combination of supervised and unsupervised learning [9]. A supervised learning machine typically yields a generalization function [math]\hat{h}(\cdot)[/math] that provides the solution to a classification problem. The main goal of classification is to categorize objects into a pre-defined set of classes. For the remainder of this section we will consider the most common classification problem, where the outcome variable [math]Y[/math] has two classes, coded as [math]\{-1,1\}[/math]. Note that this coding differs from the standard [math]\{0,1\}[/math] which is typically used in statistics for dichotomous outcomes.
The machine should learn from a training sample [math](y_1, \bm{x}_1), ..., (y_n, \bm{x}_n)[/math] with known class labels how to predict the class of a new observation [math]\bm{x}_{\text{new}}[/math]. The predictors [math]\bm{x}_1,...,\bm{x}_n[/math] are realizations of [math]X[/math], and [math]n[/math] is the sample size. The task for the machine is to develop a prediction rule [math]\hat{h}(\cdot)[/math] to correctly classify a new observation:
The success story of boosting began with a question, not with an algorithm. The theoretical discussion was if any weak learning tool for classification could be transformed to become also a strong learner [10]. In binary classification, a weak learner is defined to yield a correct classification rate at least slightly better than random guessing (> 50%). A strong learner, on the other hand, should be able to be trained to a nearly perfect classification (e.g., 99% accuracy). This theoretical question is of high practical relevance as it is typically easy to construct a weak learner, but difficult to get a strong one [11]. The answer, which laid the ground for the concept of boosting, is that any weak base-learner can be potentially iteratively improved (boosted) to become also a strong learner. To provide evidence for this concept, Schapire [12] and Freund [13] developed the first boosting algorithms.
Schapire and Freund later compared the general concept of boosting with “garnering wisdom from a council of fools” [14]. The “fools” in this case are the solutions of the simple base-learner: It classifies only slightly better than the flip of a coin. A simple base-learner is by no means a practical classification rule, but even the simple base-learner must contain some valid information about the underlying structure of the problem. The task of a boosting algorithm is hence to learn from the iterative application of a weak learner and to use this information to combine it to an accurate classification.
However, just calling the weak learner multiple times on the same training sample would not change anything in its performance. The concept of boosting is not really to manipulate the base-learner itself to improve its performance but to manipulate the underlying training data by iteratively re-weighting the observations [14]. As a result, the base-learner in every iteration [math]m[/math] will find a new solution [math]\hat{h}^{[m]}(\cdot)[/math] from the data.
Via repeated application of the weak base-learner on observations that are weighted based on the base-learner's success in the previous rounds, the algorithm is forced to concentrate on objects that are hard to classify -- as observations that were misclassified before get higher weights. Boosting the accuracy is achieved by increasing the importance of “difficult” observations. In each iteration [math]m = 1,...,m_{\text{stop}}[/math], the weight vector [math]\bm{w}^{[m]} = (w_1^{[m]} ,...,w_n^{[m]})[/math] contains the individual weights of all observations depending on the success of their classification in previous iterations. During the iteration cycle, the focus is shifted towards observations that were misclassified up to the current iteration [math]m[/math].
In a final step, all previous results of the base-learner are combined into a more accurate prediction: The weights of better performing solutions of the base-learner are increased via an iteration-specific coefficient, which depends on the corresponding misclassification rate. The resulting weighted majority vote [15] chooses the class most often selected by the base-learner while taking the error rate in each iteration into account (see point (5) in Box).
This combination of forcing the algorithm to develop new strategies for problematic observations and rewarding the base-learner in the final aggregation for accurate solutions is the main idea of boosting. Following this concept, it can be shown that all weak learners can potentially be boosted to become also strong learners [12][13].
AdaBoost
The early boosting algorithms by Schapire [12] and Freund [13] were rather theoretical constructs for proving the idea of boosting than being suitable algorithms for practical usage. However, they paved the way for the first concrete and -- still today -- most important boosting algorithm AdaBoost [7]. AdaBoost was the first adaptive boosting algorithm as it automatically adjusts its parameters to the data based on the actual performance in the current iteration: both the weights [math]w_i[/math] for re-weighting the data as well as the weights [math]\alpha_m[/math] for the final aggregation are re-computed iteratively. For a schematic overview, see Box -- for worked out examples, we refer to [11][14].
The introduction of AdaBoost gained much attention in the machine learning community. In practice, it is often used with simple classification trees or stumps as base-learners and typically results in a dramatically improved performance compared to the classification by one tree or any other single base-learner [16][17]. For example, Bauer and Kohavi [18] report an average 27\% relative improvement in the misclassification error for AdaBoost compared with a single decision tree. The authors additionally compared the accuracy of AdaBoost with the one of Bagging [19] in various settings. Bagging, in contrast to boosting, uses bootstrap generated samples to modify the training data and hence does not rely on the misclassification rate of earlier iterations. After their large-scale comparison, Bauer and Kohavi concluded that boosting algorithms, in contrast to Bagging, are able to reduce not only the variation in the base-learner's prediction error resulting from the use of different training data sets (variance), but also the average difference between predicted and true classes (bias). This view is also essentially supported by an analysis of Breiman [20]. The success of AdaBoost allegedly led Breiman, who was a pioneer and leading expert in machine learning, to the statement [21]: Boosting is the best off-the-shelf classifier in the world.
Initialization
- (1) Set the iteration counter [math]m=0[/math] and the individual weights [math]w_i[/math] for observations [math]i = 1,...,n[/math] to [math]w_i^{[0]} = \frac{1}{n}[/math].
Base-learner
- (2) Set [math]m := m + 1[/math] and compute the base-learner for the weighted data set:
[[math]] \text{re-weight observations with } w_1^{[m-1]},..., w_n^{[m-1]} \xrightarrow{\text{base-learner}} \hat{h}^{[m]}(\cdot) [[/math]]
Update weights
- (3) Compute error rate and update the iteration-specific coefficient [math]\alpha_m[/math] [math]\rightarrow[/math] high values for small error rates. Update individual weights [math]w_i^{[m]}[/math] [math]\rightarrow[/math] higher values if observation was misclassified.
Iterate
- (4) Iterate steps 2 and 3 until [math]m = m_{\text{stop}}[/math].
Final aggregation
- (5) Compute the final classifier for a new observation [math]\bm{x}_{\text{new}}[/math]:
[[math]] \hat{f}_{\text{Adaboost}}(\bm{x}_{\text{new}}) = \text{sign} \left(\sum_{m=1}^{m_{\text{stop}}} \alpha_{m} \hat{h}^{[m]}(\bm{x}_{\text{new}}) \right) [[/math]]
Overfitting
A long-lasting discussion in the context of AdaBoost is its overfitting behavior. Overfitting describes the common phenomenon that when a prediction rule concentrates too much on peculiarities of the specific sample of training observations it was optimized on, it will often perform poorly on a new data set [22]. To avoid overfitting, the task for the algorithm therefore should not be to find the best possible classifier for the underlying training sample, but rather to find the best prediction rule for a set of new observations.
The main control instrument to avoid overfitting in boosting algorithms is the stopping iteration [math]m_{\text{stop}}[/math]. Very late stopping of AdaBoost may favor overfitting, as the complexity of the final solution increases. On the other hand, stopping the algorithm too early does not only inevitably lead to higher error on the training data but could as well result in a poorer prediction on new data (underfitting). In the context of AdaBoost, it is nowadays consensus that although the algorithm may overfit [23][24] , it often is quite resistent to overfitting [11][14].
In their initial article, Freund and Schapire [7] showed that the generalization error on a test data set of AdaBoost's final solution is bounded by the training error plus a term which increases with the number of boosting iterations and the complexity of the base-learner. This finding was apparently supported by the widely acknowledged principle known as Occam's Razor [25], which roughly states that for predictions, more complex classifiers should be outperformed by less complex ones if both carry the same amount of information. However, this theoretical result is not supported by the observation that AdaBoost, in practice, is often resistent to overfitting. As the complexity of the final AdaBoost solution depends mainly on the stopping iteration [math]m_{\text{stop}}[/math], following Occam's Razor, later stopping of the algorithm should yield poorer predictions [11].
One way to explain AdaBoost's overfitting behavior is based on the margin interpretation [17][24][26]: The margin of the final boosting solution, in brief, can be interpreted as the confidence in the prediction. With higher values of [math]m_{\text{stop}}[/math], this margin may still increase and lead to better predictions on the test data even if the training error is already zero [27]. This theory was early questioned by results of Breiman [28], who developed the arc-gv algorithm which should yield a higher margin than AdaBoost, but clearly failed to outperform it in practice with respect to prediction accuracy. Later, Reyzin and Schapire [27] explained these findings with other factors like the complexity of the base-learner. For more on the margin interpretation see the corresponding chapters in [11][14].
Another explanation of the -- seemingly contradictory -- results on the overfitting behavior of boosting is the use of the wrong performance criteria for evaluation (e.g.,[29]). The performance of AdaBoost has often been measured by evaluating the {\em correct classification rate}, and the resistance to overfitting has usually been demonstrated by focusing on this specific criterion only. However, the criterion that is optimized by AdaBoost is in fact not the correct classification rate but the so-called {\em exponential loss function}, and it can be shown that the two criteria are not necessarily optimized by the same predictions. For this reason some authors have argued that the overfitting behavior of AdaBoost should be analyzed by solely focusing on the exponential loss function [30]. For example, Bühlmann and Yu [31] have provided empirical evidence that too large [math]m_{\text{stop}}[/math] can lead to overfitting regarding the exponential loss without affecting the misclassification rate.
Statistical boosting
Up to this point, we focused on the classical supervised learning problem where the task of boosting is to predict dichotomous outcomes. Nowadays, boosting algorithms are more often used to estimate the unknown quantities in general statistical models (statistical boosting). In the remainder of this section, we will therefore broaden the scope and consider general regression settings where the outcome variable [math]Y[/math] can also be continuous or represent count data. The most important interpretation of boosting in this context is the statistical view of boosting by Friedman et al.[8]. It provided the basis for understanding the boosting concept in general and the success of AdaBoost in particular from a statistical point of view [16] by showing that AdaBoost in fact fits an additive model.
Most solutions of machine-learning algorithms, including AdaBoost, must be seen as black-box prediction schemes. They might yield very accurate predictions for future or unobserved data, but the way those results are produced and which role single predictors play are hardly interpretable. A statistical model, in contrast, aims at quantifying the relation between one or more observed predictor variables [math]\bm{x}[/math] and the expectation of the response [math]\text{E}(Y)[/math] via an interpretable function [math]\text{E}(Y|X=\bm{x}) = f(\bm{x})[/math]. In cases of more than one predictor, the different effects of the single predictors are typically added, forming an additive model
where [math]\beta_0[/math] is an intercept and [math]h_1(\cdot)[/math],...,[math]h_p(\cdot)[/math] incorporate the effects of predictors [math]x_1, ..., x_p[/math], which are components of [math]X[/math]. The corresponding model class is called generalized additive models ('GAM', [32]) and the aim is to model the expected value of the response variable, given the observed predictors via a link-function [math]g(\cdot)[/math]:
GAMs are by definition no black boxes but contain interpretable additive predictors: The partial effect of predictor [math]x_1[/math], for example, is represented by [math]h_1(\cdot)[/math]. The direction, the size and the shape of the effect can be visualized and interpreted -- this is a main difference towards many tree-based machine learning approaches.
The core message delivered with the statistical view of boosting is that the original AdaBoost algorithm with regression-type base-learners (e.g., linear models, smoothing splines), in fact, fits a GAM for dichotomous outcomes via the exponential loss in a stage-wise manner. The work by Friedman et al.[8] therefore provided the link between a successful machine-learning approach and the world of statistical modelling [16].
Gradient boosting
The concept of the statistical view of boosting was further elaborated by Friedman [33] who presented a boosting algorithm optimizing the empirical risk via steepest gradient descent in function space. Generally, the optimization problem for estimating the regression function [math]f(\cdot)[/math] of a statistical model, relating the predictor variables [math]X[/math] with the outcome [math]Y[/math], can be expressed as
where [math]\rho(\cdot)[/math] denotes a loss function. The most common loss function is the [math]L_2[/math] loss [math]\rho(y,f(\cdot)) = (y - f(\cdot))^2[/math], leading to classical least squares regression of the mean: [math]f(\bm{x}) = \text{E} (Y | X = \bm{x})[/math]. In practice, with a learning sample of observations [math](y_1, \bm{x}_1), ...,(y_n, \bm{x}_n)[/math] we minimize the empirical risk:
The fundamental idea of gradient boosting is to fit the base-learner not to re-weighted observations, as in AdaBoost, but to the negative gradient vector [math]\bm{u}^{[m]}[/math] of the loss function [math]\rho(y, \hat{f}(x))[/math] evaluated at the previous iteration [math]m-1[/math]:
In case of the [math]L_2[/math] loss, [math]\rho(y,f(\cdot)) = \frac{1}{2} (y - f(\cdot))^2[/math] leads simply to re-fitting the residuals [math]y - f(\cdot)[/math]. In every boosting iteration [math]m[/math], the base-learner is hence directly fitting the errors made in the previous iteration [math]y - f(\cdot)^{[m-1]}[/math]. Keeping this principle in mind, it becomes obvious that both AdaBoost and gradient boosting follow the same fundamental idea: Both algorithms boost the performance of a simple base-learner by iteratively shifting the focus towards problematic observations that are ‘difficult’ to predict. With AdaBoost, this shift is done by up-weighting observations that were misclassified before. Gradient boosting identifies difficult observations by large residuals computed in the previous iterations.
Initialization
- (1) Set the iteration counter [math]m=0[/math]. Initialize the additive predictor [math]\hat{f}^{[0]}[/math] with a starting value, e.g. [math]\hat{f}^{[0]} := (\bm{0})_{i=1,...,n}[/math]. Specify a set of base-learners [math]h_1(x_1),..., h_p(x_p)[/math].
Fit the negative gradient
- Set [math]m := m + 1[/math].
- (2) Compute the negative gradient vector [math]\bm{u}[/math] of the loss function evaluated at the previous iteration:
[[math]] \bm{u}^{[m]} = \left( u^{[m]}_i \right)_{i=1,...,n} = \left( - \left. \frac{\partial }{\partial f} \rho(y_i,f )\right|_{ f = \hat{f}^{[m-1]}(\cdot)} \right)_{i=1,...,n} [[/math]]
- (3) Fit the negative gradient vector [math]\bm{u}^{[m]}[/math] separately to every base-learner:
[[math]] \bm{u}^{[m]} \xrightarrow{\rm base-learner} \hat{h}_j^{[m]}(x_j) \quad \text{for } j=1,...,p. [[/math]]
Update one component
- (4) Select the component [math]j^*[/math] that best fits the negative gradient vector:
[[math]] j^* = \underset{1 \leq j \leq p}{\operatorname{argmin}}\sum_{i=1}^n (u_{i}^{[m]} - \hat{h}_{j}^{[m]}(x_j))^2 \ . [[/math]]
- (5) Update the additive predictor [math]\hat{f}[/math] with this component
[[math]] \hat{f}^{[m]}(\cdot) = \hat{f}^{[m-1]} (\cdot) + \text{sl} \cdot \hat{h}_{j^*}^{[m]}(x_{j^*}) \ , [[/math]]where sl is a small step length [math](0 \lt \text{sl} \ll 1)[/math]. A typical value in practice is 0.1.
Iteration
Iterate steps (2) to (6) until [math]m=m_{\text{stop}}[/math]
Generally, the underlying base-learner can be any regression technique; the most simple base-learner is a classical linear least-squares model with [math]h(\bm{x}) = \bm{x}^{\top}\beta[/math]. If [math]\bm{x}[/math] is assumed to have a non-linear effect on the response, smoothing splines could be used [33]. Bühlmann and Yu [34] further developed the gradient boosting approach by applying component-wise smoothing splines as base-learners. The fundamental idea is that different predictors are fitted by separate base-learners [math]h_j(\cdot)[/math], [math]j=1,...,p[/math]. Typically, each base-learner [math]h_j(\cdot)[/math] corresponds to one component [math]x_j[/math] of [math]X[/math] and in every boosting iteration (as proposed in [33]) only a small amount of the fit of the best-performing base-learner is added to the current additive predictor. The authors demonstrated that the resulting algorithm in combination with the [math]L_2[/math] loss outperforms classical additive modelling in terms of prediction accuracy. This approach was further developed by Bühlmann [35] who specially focused on high-dimensional data settings.
Bühlmann and Hothorn [36] gave an overview of gradient boosting algorithms from a statistical perspective presenting a generic functional gradient descent algorithm (see Box). As in [33], base-learners are used to fit the negative gradient vector of the corresponding loss function. The algorithm descends the empirical risk via steepest gradient descent in function space, where the function space is provided by the base-learners. Each base-learner typically includes one predictor and in every boosting iteration only the best-performing base-learner and hence the best performing component of [math]X[/math] is included in the final model. This procedure effectively leads to data-driven variable selection during the model estimation. The base-learners [math]h_1(x_1),..., h_p(x_p)[/math] reflect the type of effect the corresponding components will contribute to the final additive model, which offers the same interpretability as any other additive modelling approach. Examples for base-learners can be trees as in classical boosting algorithms, but commonly simple regression tools like linear models or splines are used to include linear as well as non-linear effects on the response. Generally, it is consensus in the literature that base-learners should be weak in the sense that they do not offer too complex solutions in a single iteration (e.g., penalized splines with small degrees of freedom [37]). However, there is no formal definition for weak or strong learners in the context of statistical boosting.
In contrast to standard estimation methods, component-wise gradient boosting also works for high dimensional data where the number of predictors exceeds the number of observations ([math]p \gt n[/math]). Furthermore, it is relatively robust in cases of multicollinearity. Due to the small step length in the update step (a typical value is 0.1 [38]) in combination with early stopping (Section 3.3), gradient boosting incorporates shrinkage of effect estimates in the estimation process: The absolute size of the estimated coefficients is intentionally reduced -- this is a similarity to penalized regression approaches as the Lasso [39]. Shrinkage of effect estimates leads to a reduced variance of estimates and should therefore increase the stability and accuracy of predictions [21].
Early stopping of statistical boosting algorithms
Although there are different influential factors for the performance of boosting algorithms, the stopping iteration [math]m_{\text{stop}}[/math] is considered to be the main tuning parameter [40]. Stopping the algorithm before its convergence (early stopping) prevents overfitting (Section 2.3) and typically improves prediction accuracy. In case of statistical boosting, [math]m_{\text{stop}}[/math] controls both shrinkage of effect estimates and variable selection. The selection of [math]m_{\text{stop}}[/math] hence reflects the common bias-variance trade-off in statistical modelling: Large values of [math]m_{\text{stop}}[/math] lead to more complex models with higher variance and small bias. Smaller values of [math]m_{\text{stop}}[/math] lead to sparser models with less selected variables, more shrinkage and reduced variance [40].
To prevent overfitting, it is crucial not to consider the stopping iteration [math]m_{\text{stop}}[/math] that leads to the best model on the training data but to evaluate the effect of [math]m_{\text{stop}}[/math] on separate test data. If no additional data are available, two general approaches are commonly applied:
The first is to use information criteria (AIC, BIC or gMDL [41]) which evaluate the likelihood on training data but additionally penalize too complex models by adding a multiple of their degrees of freedom. There are two problems with this approach: (i) for component-wise boosting algorithms these information criteria rely on an estimation of the degrees of freedom that is known to underestimate the true values [42]; (ii) they are only available for a limited number of loss functions.
The second, more general approach is to apply resampling or cross-validation techniques to subsequently divide the data into test and training sets and choose [math]m_{\text{stop}}[/math] by evaluating the models on the test data. For the evaluation, it is crucial to use the same loss function the algorithm aims to optimize. If the algorithm in a binary classification setting optimizes the exponential loss, one should use the exponential loss and not the misclassification rate to select [math]m_{\text{stop}}[/math]. The optimal [math]m_{\text{stop}}[/math] is hence the one which leads to the smallest average empirical loss on the out-of-sample test data.
Random Forests
This section is based on [43]. To take advantage of the sheer size of modern data sets, we now need learning algorithms that scale with the volume of information, while maintaining sufficient statistical efficiency. Random forests, devised by L. Breiman in the early 2000s [44],are part of the list of the most successful methods currently available to handle data in these cases. This supervised learning procedure, influenced by the early work of [45], [46], and [47], operates according to the simple but effective divide and conquer principle: sample fractions of the data, grow a randomized tree predictor on each small piece, then paste (aggregate) these predictors together.
What has greatly contributed to the popularity of forests is the fact that they can be applied to a wide range of prediction problems and have few parameters to tune. Aside from being simple to use, the method is generally recognized for its accuracy and its ability to deal with small sample sizes and high-dimensional feature spaces. At the same time, it is easily parallelizable and has therefore the potential to deal with large real-life systems.
Basic Principles
As mentioned above, the forest mechanism is versatile enough to deal with both supervised classification and regression tasks. However, to keep things simple, we focus in this introduction on regression analysis, and only briefly survey the classification case. Our objective in this section is to provide a concise but mathematically precise presentation of the algorithm for building a random forest. The general framework is nonparametric regression estimation, in which an input random vector [math]\bX \in \mathcal{X} \subset \mathbb{R}^p[/math] is observed, and the goal is to predict the square integrable random response [math]Y\in \mathbb R[/math] by estimating the regression function [math]m(\bx)=\mathbb E[Y|\bX=\bx][/math]. With this aim in mind, we assume we are given a training sample [math]\mathcal D_n=((\bX_1,Y_1), \ldots, (\bX_n,Y_n))[/math] of independent random variables distributed as the independent prototype pair [math](\bX, Y)[/math]. The goal is to use the data set [math]\mathcal D_n[/math] to construct an estimate [math]m_n: \mathcal{X} \to \mathbb R[/math] of the function [math]m[/math]. In this respect, we say that the regression function estimate [math]m_n[/math] is (mean squared error) consistent if [math]\mathbb E [m_n(\bX)-m(\bX)]^2 \to 0[/math] as [math]n \to \infty[/math] (the expectation is evaluated over [math]\bX[/math] and the sample [math]\mathcal D_n[/math]).
A random forest is a predictor consisting of a collection of [math]M[/math] randomized regression trees. For the [math]j[/math]-th tree in the family, the predicted value at the query point [math]\bx[/math] is denoted by [math]m_n(\bx; \Theta_j,\mathcal D_n)[/math], where [math]\Theta_1, \ldots,\Theta_M[/math] are independent random variables, distributed the same as a generic random variable [math]\Theta[/math] and independent of [math]\mathcal D_n[/math]. In practice, the variable [math]\Theta[/math] is used to resample the training set prior to the growing of individual trees and to select the successive directions for splitting---more precise definitions will be given later. In mathematical terms, the [math]j[/math]-th tree estimate takes the form
where [math]\mathcal{D}_n^{\star}(\Theta_j)[/math] is the set of data points selected prior to the tree construction, [math]A_n(\bx; \Theta_j, \mathcal{D}_n)[/math] is the cell containing [math]\bx[/math], and [math]N_n(\bx; \Theta_j, \mathcal{D}_n)[/math] is the number of (preselected) points that fall into [math]A_n(\bx; \Theta_j, \mathcal{D}_n)[/math].
At this stage, we note that the trees are combined to form the (finite) forest estimate
In the R package randomForest, the default value of [math]M[/math] (the number of trees in the forest) is ntree = 500. Since [math]M[/math] may be chosen arbitrarily large (limited only by available computing resources), it makes sense, from a modeling point of view, to let [math]M[/math] tends to infinity, and consider instead of (\ref{chapitre0_finite_forest}) the (infinite) forest estimate \begin{align*} m_{\infty, n}(\bx; \mathcal{D}_n)=\mathbb E_{\Theta}\left [m_n(\bx;\Theta,\mathcal D_n)\right]. \end{align*} In this definition, [math]\mathbb E_{\Theta}[/math] denotes the expectation with respect to the random parameter [math]\Theta[/math], conditional on [math]\mathcal D_n[/math]. In fact, the operation [math]M \to \infty[/math] is justified by the law of large numbers, which asserts that almost surely, conditional on [math]\mathcal{D}_n[/math],
(see for instance [44], and [48], for more information on this limit calculation). In the following, to lighten notation we will simply write [math]m_{\infty, n}(\bx)[/math] instead of [math]m_{\infty, n}(\bx;\mathcal{D}_n)[/math].
Algorithm
We now provide some insight on how the individual trees are constructed and how randomness kicks in. In Breiman’s [44] original forests, each node of a single tree is associated with a hyperrectangular cell. The root of the tree is [math]\mathcal{X}[/math] itself and, at each step of the tree construction, a node (or equivalently its corresponding cell) is split in two parts. The terminal nodes (or leaves), taken together, form a partition of [math]\mathcal{X}[/math].
The algorithm works by growing [math]M[/math] different (randomized) trees as follows. Prior to the construction of each tree, [math]a_n[/math] observations are drawn at random with (or without) replacement from the original data set. These---and only these---[math]a_n[/math] observations (with possible repetitions) are taken into account in the tree building. Then, at each cell of each tree, a split is performed by maximizing the CART-criterion (see below) over mtry directions chosen uniformly at random among the [math]p[/math] original ones. (The resulting subset of selected coordinates is called mtry.) Lastly, construction of individual trees is stopped when each cell contains less than nodesize points. For any query point [math]\bx \in \mathcal X[/math], each regression tree predicts the average of the [math]Y_i[/math] (that were among the [math]a_n[/math] points) for which the corresponding [math]\bX_i[/math] falls into the cell of [math]\bx[/math]. We draw attention to the fact that growing the tree and making the final estimation only depends on the [math]a_n[/math] preselected data points. Algorithm 1 describes in full detail how to compute a forest's prediction.
Input: Training set [math]\mathcal{D}_n[/math], number of trees [math]M\gt0[/math], [math]a_n \in \{1, \ldots, n \}[/math], [math]\texttt{mtry} \in \{1, \ldots, p \}[/math], [math]\texttt{nodesize} \in \{1, \ldots, a_n \}[/math], and [math]\bx \in \mathcal{X}[/math].
Output: Prediction of the random forest at [math]\bx[/math].
For [math]j=1,\ldots,M[/math] do
- Select [math]a_n[/math] points, with (or without) replacement, uniformly in [math]\mathcal{D}_n[/math]. In the following steps, only these [math]a_n[/math] observations are used.
- Set [math]\mathcal{P} = (\mathcal{X})[/math] the list containing the cell associated with the root of the tree.
- Set [math]\Pfin = \emptyset[/math] an empty list.
While: [math]\mathcal{P} \neq \emptyset [/math] do
Let [math]A[/math] be the first element of [math]\mathcal{P}[/math]
If [math]A[/math] contains less than nodesize points or if all [math]\bX_i \in A[/math] are equal
then
- Remove the cell [math]A[/math] from the list [math]\mathcal{P}[/math].
- [math]\Pfin \leftarrow Concatenate (\Pfin , A)[/math].
else
- Select uniformly, without replacement, a subset [math]\Mtry \subset[/math] [math] \{1,\ldots,[/math] [math]p\}[/math] of cardinality mtry.
- Select the best split in [math]A[/math] by optimizing the CART-split criterion along the coordinates in [math]\mathcal{M}_{\textrm{try}}[/math] (see text for details).
- Cut the cell [math]A[/math] according to the best split. Call [math]A_L[/math] and [math]A_R[/math] the two resulting cells.
- Remove the cell [math]A[/math] from the list [math]\mathcal{P}[/math].
- [math]\mathcal{P} \leftarrow Concatenate( \mathcal{P}, A_L, A_R)[/math].
end
Compute the predicted value [math]m_n(\bx; \Theta_j, \mathcal{D}_n)[/math] at [math]\bx[/math] equal to the average of the [math]Y_i[/math] falling in the cell of [math]\bx[/math] in partition [math]\Pfin[/math].
end
Compute the random forest estimate [math]m_{M,n}(\bx; \Theta_1, \ldots, \Theta_M, \mathcal{D}_n)[/math] at the query point [math]\bx[/math] according to (\ref{chapitre0_finite_forest}).
Algorithm 1 may seem a bit complicated at first sight, but the underlying ideas are simple. We start by noticing that this algorithm has three important parameters:
- [math]a_n \in \{1, \ldots, n \}[/math]: the number of sampled data points in each tree;
- [math]\mtry \in \{1, \ldots, p \}[/math]: the number of possible directions for splitting at each node of each tree;
- [math]\texttt{nodesize} \in \{1, \ldots, a_n \}[/math]: the number of examples in each cell below which the cell is not split.
By default, in the regression mode of the R package randomForest, the parameter mtry is set to [math]\lceil p/3 \rceil[/math] ([math]\lceil \cdot \rceil[/math] is the ceiling function), [math]a_n[/math] is set to [math]n[/math], and nodesize is set to [math]5[/math]. The role and influence of these three parameters on the accuracy of the method will be thoroughly discussed in the next section.
We still have to describe how the CART-split criterion operates. As for now, we consider for the ease of understanding a tree with no subsampling, which uses the entire and original data set [math]\mathcal{D}_n[/math] for its construction. Also, we let [math]A[/math] be a generic cell and denote by [math]N_n(A)[/math] the number of data points falling in [math]A[/math]. A cut in [math]A[/math] is a pair [math](j,z)[/math], where [math]j[/math] is some value (dimension) from [math]\{1, \ldots, p\}[/math] and [math]z[/math] the position of the cut along the [math]j[/math]-th coordinate, within the limits of [math]A[/math]. Let [math]\mathcal{C}_A[/math] be the set of all such possible cuts in [math]A[/math]. Then, with the notation [math]\bX_i = (\bX_i^{(1)}, \ldots, \bX_i^{(p)} )[/math], for any [math](j,z) \in \mathcal{C}_A[/math], the CART-split criterion takes the form
where [math]A_L = \{ \bx \in A: \bx^{(j)} \lt z\}[/math], [math]A_R = \{ \bx \in A: \bx^{(j)} \geq z\}[/math], and [math]\bar{Y}_{A}[/math] (resp., [math]\bar{Y}_{A_{L}}[/math], [math]\bar{Y}_{A_{R}}[/math]) is the average of the [math]Y_i[/math] belonging to [math]A[/math] (resp., [math]A_{L}[/math], [math]A_{R}[/math]), with the convention that the average is equal to [math]0[/math] when no point [math]\bX_i[/math] belongs to [math]A[/math] (resp., [math]A_{L}[/math], [math]A_{R}[/math]). For each cell [math]A[/math], the best cut [math](j_n^{\star},z_n^{\star})[/math] is selected by maximizing [math]L_{\textrm{reg}, n}(j,z)[/math] over mtry and [math]\mathcal{C}_A[/math]; that is,
(To remove some of the ties in the argmax, the best cut is always performed in the middle of two consecutive data points.) Let us finally notice that the above optimization program extends effortlessly to the resampling case, by optimizing over the [math]a_n[/math] preselected observations instead of the original data set [math]\mathcal{D}_n[/math].
Thus, at each cell of each tree, the algorithm chooses uniformly at random mtry coordinates in [math]\{1, \ldots, p\}[/math], evaluates criterion (\ref{chapitre0_definition_empirical_CART_criterion}) over all possible cuts along the directions in mtry, and returns the best one. The quality measure (\ref{chapitre0_definition_empirical_CART_criterion}) is the criterion used in the influential CART algorithm of [49]. This criterion computes the (renormalized) difference between the empirical variance in the node before and after a cut is performed. There are three essential differences between CART and a tree of Breiman's [44] forest. First of all, in Breiman's forests, the criterion (\ref{chapitre0_definition_empirical_CART_criterion}) is evaluated over a subset mtry of randomly selected coordinates, and not over the whole range [math]\{1, \ldots, p\}[/math]. Besides, the individual trees are not pruned, and the final cells do not contain more than nodesize observations (unless all data points in the cell have the same [math]\bX_i[/math]). At last, each tree is constructed on a subset of [math]a_n[/math] examples picked within the initial sample, not on the whole sample [math]\mathcal D_n[/math]; and only these [math]a_n[/math] observations are used to calculate the estimation. When [math]a_n=n[/math] (and the resampling is done with replacement), the algorithm runs in bootstrap mode, whereas [math]a_n \lt n[/math] corresponds to subsampling (with or without replacement).
Supervised classification
For simplicity, we only consider here the binary classification problem, keeping in mind that random forests are intrinsically capable of dealing with multi-class problems [50]. In this setting [51], the random response [math]Y[/math] takes values in [math]\{0, 1\}[/math] and, given [math]\bX[/math], one has to guess the value of [math]Y[/math]. A classifier, or classification rule, [math]m_n[/math] is a Borel measurable function of [math]\bX[/math] and [math]\mathcal D_n[/math] that attempts to estimate the label [math]Y[/math] from [math]\bX[/math] and [math]\mathcal D_n[/math]. In this framework, one says that the classifier [math]m_n[/math] is consistent if its conditional probability of error
where [math]L^{\star}[/math] is the error of the optimal---but unknown---Bayes classifier:
In the classification context, the random forest classifier is obtained via a majority vote among the classification trees, that is,
If a leaf represents region [math]A[/math], then a randomized tree classifier takes the simple form
where [math]\mathcal{D}^{\star}_n(\Theta)[/math] contains the data points selected in the resampling step. That is, in each leaf, a majority vote is taken over all [math](\bX_i,Y_i)[/math] for which [math]\bX_i[/math] is in the same region. Ties are broken, by convention, in favor of class 0. Algorithm 1 can be easily adapted to do classification by modifying the CART-split criterion for the binary setting. To see this, let us consider a single tree with no subsampling step. For any generic cell [math]A[/math], let [math]p_{0,n}(A)[/math] (resp., [math]p_{1,n}(A)[/math]) be the empirical probability of a data point in the cell [math]A[/math] having label [math]0[/math] (resp., label [math]1[/math]). Then, for any [math](j,z) \in \mathcal{C}_A[/math], the classification CART-split criterion reads
This criterion is based on the so-called Gini impurity measure [math]2p_{0,n}(A)p_{1,n}(A)[/math] [49], which has the following simple interpretation. To classify a data point that falls in cell [math]A[/math], one uses the rule that assigns a point, uniformly selected from [math]\{\bX_i \in A: (\bX_i, Y_i)\in \mathcal{D}_n\}[/math], to label [math]\ell[/math] with probability [math]p_{\ell,n}(A)[/math], for [math]j \in \{0,1\}[/math]. The estimated probability that the item has actually label [math]\ell[/math] is [math]p_{\ell,n}(A)[/math]. Therefore the estimated error under this rule is the Gini index [math]2p_{0,n}(A)p_{1,n}(A)[/math].
We note that whenever [math]Y \in \{0,1\}[/math], optimizing the classification criterion [math]L_{\textrm{class}, n}[/math] is equivalent to optimizing its regression counterpart [math]L_{\textrm{reg}, n}[/math]. Thus, in this case, the trees obtained with [math]L_{\textrm{class}, n}[/math] and [math]L_{\textrm{reg}, n}[/math] are identical. However, the prediction strategy is different: in the classification regime, each tree uses a local majority vote, whereas in regression the prediction is achieved by a local averaging.
When dealing with classification problems, it is usually recommended to set nodesize = 1 and mtry=[math]\sqrt{p}[/math] [52].
We draw attention to the fact that regression estimation may also have an interest in the context of dichotomous and multicategory outcome variables (in this case, it is often termed probability estimation). For example, estimating outcome probabilities for individuals is important in many areas of medicine, with applications to surgery, oncology, internal medicine, pathology, pediatrics, and human genetics. We refer the interested reader to [53] and to the survey papers by [54] and [55].
Parameter tuning
Literature focusing on tuning the parameters [math]M[/math], mtry, nodesize and [math]a_n[/math] is unfortunately rare, with the notable exception of [50], [56], and [57]. According to [58], tuning the forest parameters may result in a computational burden, in particular for big data sets, with hundreds and thousands of samples and variables. To circumvent this issue, [58] implement a fast version of the original algorithm, which they name Random Jungle.
It is easy to see that the forest's variance decreases as [math]M[/math] grows. Thus, more accurate predictions are likely to be obtained by choosing a large number of trees. Interestingly, picking a large [math]M[/math] does not lead to overfitting. In effect, following an argument of [44], we have
However, the computational cost for inducing a forest increases linearly with [math]M[/math], so a good choice results from a trade-off between computational complexity ([math]M[/math] should not be too large for the computations to finish in a reasonable time) and accuracy ([math]M[/math] must be large enough for predictions to be stable). In this respect, [50] argue that the value of [math]M[/math] is irrelevant (provided that [math]M[/math] is large enough) in a prediction problem involving microarray data sets, where the aim is to classify patients according to their genetic profiles (typically, less than one hundred patients for several thousand genes). For more details we refer the reader to [57], who offer a thorough discussion on the choice of this parameter in various regression problems. Another interesting and related approach is by [59], who propose a simple procedure that determines a priori a minimum number of tree estimates to combine in order to obtain a prediction accuracy level similar to that of a larger forest. Their experimental results show that it is possible to significantly limit the number of trees.
In the R package randomForest, the default value of the parameter nodesize is 1 for classification and 5 for regression. These values are often reported to be good choices [50] despite the fact that this is not supported by solid theory. A simple algorithm to tune the parameter nodesize in the classification setting is discussed in [60].
The effect of mtry is thoroughly investigated in [50], who show that this parameter has a little impact on the performance of the method, though larger values may be associated with a reduction in the predictive performance. On the other hand, [57] claim that the default value of mtry is either optimal or too small. Therefore, a conservative approach is to take mtry as large as possible (limited by available computing resources) and set [math]\mtry=p[/math] (recall that [math]p[/math] is the dimension of the [math]\bX_i[/math]). A data-driven choice of mtry is implemented in the algorithm Forest-RK of [56].
Let us finally notice that even if there is no theoretical guarantee to support the default values of the parameters, they are nevertheless easy to tune without requiring an independent validation set. Indeed, the procedure accuracy is estimated internally, during the run, as follows. Since each tree is constructed using a different bootstrap sample from the original data, about one-third of the observations are left out of the bootstrap sample and not used in the construction of the [math]j[/math]-th tree. In this way, for each tree, a test set---disjoint from the training set---is obtained, and averaging over all these left-out data points and over all trees is known as the out-of-bag error estimate. Thus, the out-of-bag error, computed on the observations set aside by the resampling prior to the tree building, offers a simple way to adjust the parameters without the need of a validation set. (e.g.,[60]).
Variable importance measures
Random forests can be used to rank the importance of variables in regression or classification problems via two measures of significance. The first, called Mean Decrease Impurity (see [61]), is based on the total decrease in node impurity from splitting on the variable, averaged over all trees. The second, referred to as Mean Decrease Accuracy (MDA), first defined by [44], stems from the idea that if the variable is not important, then rearranging its values should not degrade prediction accuracy.
Set [math]\bX = (X^{(1)}, \ldots, X^{(p)} )[/math]. For a forest resulting from the aggregation of [math]M[/math] trees, the MDI of the variable [math]X^{(j)}[/math] is defined by
where [math]p_{n,t}[/math] is the fraction of observations falling in the node [math]t[/math], [math]\{\mathcal{T}_{\ell}\}_{1\leq {\ell} \leq M}[/math] the collection of trees in the forest, and [math](j_{n,t}^{\star}, z_{n,t}^{\star})[/math] the split that maximizes the empirical criterion (\ref{chapitre0_definition_empirical_CART_criterion}) in node [math]t[/math]. Note that the same formula holds for classification random forests by replacing the criterion [math]L_{{\scriptsize \mbox{reg}}, n}[/math] by its classification version [math]L_{{\scriptsize \mbox{class}}, n}[/math]. Thus, the MDI of [math]X^{(j)}[/math] computes the weighted decrease of impurity corresponding to splits along the variable [math]X^{(j)}[/math] and averages this quantity over all trees.
The MDA relies on a different principle and uses the out-of-bag error estimate (see Section [math]2.4[/math]). To measure the importance of the [math]j[/math]-th feature, we randomly permute the values of variable [math]X^{(j)}[/math] in the out-of-bag observations and put these examples down the tree. The MDA of [math]X^{(j)}[/math] is obtained by averaging the difference in out-of-bag error estimation before and after the permutation over all trees. In mathematical terms, consider a variable [math]X^{(j)}[/math] and denote by [math]\mathcal{D}_{{\ell},n}[/math] the out-of-bag data set of the [math]{\ell}[/math]-th tree and [math]{\mathcal{D}}^j_{{\ell},n}[/math] the same data set where the values of [math]X^{(j)}[/math] have been randomly permuted. Recall that [math]m_n(\cdot; \Theta_{\ell})[/math] stands for the [math]{\ell}[/math]-th tree estimate. Then, by definition,
where [math]R_n[/math] is defined for [math]\mathcal D=\mathcal{D}_{{\ell},n}[/math] or [math]\mathcal D={\mathcal{D}}^j_{{\ell},n}[/math] by
It is easy to see that the population version of [math]\widehat{\mbox{MDA}}(X^{(j)}) [/math] is
where [math]\bX'_{j} = (X^{(1)}, \ldots, X'^{(j)} , \ldots, X^{(p)})[/math] and [math]X'^{(j)}[/math] is an independent copy of [math]X^{(j)}[/math]. For classification purposes, the MDA still satisfies (\ref{test_bis}) and (\ref{test_bisb}) since [math]Y_i \in \{0,1\}[/math] (so, [math]R_n(m_n(\cdot; \Theta),\mathcal{D})[/math] is also the proportion of points that are correctly classified by [math]m_n(\cdot; \Theta)[/math] in [math]\mathcal{D}[/math]).
References
- Aslam, Javed A.; Popa, Raluca A.; and Rivest, Ronald L. (2007); On Estimating the Size and Confidence of a Statistical Audit, Proceedings of the Electronic Voting Technology Workshop (EVT '07), Boston, MA, August 6, 2007. More generally, when drawing with replacement [math]n[/math] values out of a set of [math]n[/math] (different and equally likely), the expected number of unique draws is [math]n(1 - e^{-n'/n})[/math].
- 2.0 2.1 Breiman, Leo (1996). "Bagging predictors". Machine Learning 24 (2): 123–140. doi:.
- Breiman, Leo (September 1994). "Bagging Predictors". Technical Report. Department of Statistics, University of California Berkeley.
- Sahu, A., Runger, G., Apley, D., Image denoising with a multi-phase kernel principal component approach and an ensemble version, IEEE Applied Imagery Pattern Recognition Workshop, pp.1-7, 2011.
- Shinde, Amit, Anshuman Sahu, Daniel Apley, and George Runger. "Preimages for Variation Patterns from Kernel PCA and Bagging." IIE Transactions, Vol.46, Iss.5, 2014
- Mayr, Andreas; Binder, Harald; Gefeller, Olaf; Schmid, Matthias (2014). "The Evolution of Boosting Algorithms From Machine Learning to Statistical Modelling". arXiv:1403.1452 [stat.ME].
- 7.0 7.1 7.2 Freund Y, Schapire R (1996). Experiments With a New Boosting Algorithm. In: Proceedings of the Thirteenth International Conference on Machine Learning Theory. San Francisco, CA: San Francisco: Morgan Kaufmann Publishers Inc. pp. 148–156.
- 8.0 8.1 8.2 Friedman JH, Hastie T, Tibshirani R. (2000). "Additive Logistic Regression: A Statistical View of Boosting (with Discussion)". The Annals of Statistics. 28: 337-407.
- Bishop, CM; et al. (2006). Pattern Recognition and Machine Learning. 4. New York: Springer.
- Kearns MJ, Valiant LG. (1989). "Cryptographic Limitations on Learning Boolean Formulae and Finite Automata". Johnson DS, editor. Proceedings of the 21st Annual ACM Symposium on Theory of Computing: 434--444.
- 11.0 11.1 11.2 11.3 11.4 Zhou, ZH. (2012). Ensemble Methods: Foundations and Algorithms. CRC Machine Learning & Pattern Recognition. Chapman & Hall.
- 12.0 12.1 12.2 Schapire RE (1990). "The Strength of Weak Learnability". Machine Learning. 5(2): 5(2)-2.
- 13.0 13.1 13.2 Freund, Y. (1990). "Boosting a Weak Learning Algorithm by Majority". In: Fulk MA, Case J, editors. Proceedings of the Third Annual Workshop on Computational Learning Theory: 202--216.
- 14.0 14.1 14.2 14.3 14.4 Schapire RE, Freund Y. (2012). Boosting: Foundations and Algorithms. MIT Press.
- Littlestone N, Warmuth MK. (1989). The Weighted Majority Algorithm. In: Foundations of Computer Science, 1989., 30th Annual Symposium on.IEEE. Springer. pp. 256--261.
- 16.0 16.1 16.2 Ridgeway G (31). "The State of Boosting". Computing Science and Statistics. 31: 172-181.
- 17.0 17.1 Meir R, Ratsch G. (2003). An Introduction to Boosting and Leveraging. Advanced Lectures on Machine Learning. Springer. pp. 118--183.
- Bauer E, Kohavi R (36). "An Empirical Comparison of Voting Classification Algorithms: Bagging, Boosting, and Variants". Journal of Machine Learning. 36: 105-139.
- Breiman L (1996). "Bagging Predictors". Machine Learning. 24: 123-140.
- Breiman L (1998). "Arcing Classifiers (with Discussion)". The Annals of Statistics. 26: 26-801.
- 21.0 21.1 Hastie T, Tibshirani R, Friedman J. (2009). The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York: Springer.CS1 maint: multiple names: authors list (link)
- Dietterich T (1995). "Overfitting and Undercomputing in Machine Learning". ACM Computing Surveys (CSUR). 27(3): 326-327.
- Grove AJ, Schuurmans D. (1998). Boosting in the Limit: Maximizing the Margin of Learned Ensembles. In: Proceeding of the AAAI-98 Learning. John Wiley & Sons Ltd. pp. 692--699.
- 24.0 24.1 Ratsch G, Onoda T, Muller KR (2001). "Soft Margins for AdaBoost". Machine Learning. 42(3): 287-320.
- Blumer A, Ehrenfeucht A, Haussler D, Warmuth MK (1987). "Occam's Razor". Information Processing Letters. 24: 377-380.
- Schapire RE, Freund Y, Bartlett P, Lee WS (1998). "Boosting The Margin: A new Explanation for the Effectiveness of Voting Methods". The Annals of Statistics. 26(5): 1651-1686.
- 27.0 27.1 Reyzin L, Schapire RE. (2003). How Boosting the Margin can also Boost Classifier Complexity. Proceeding of the 23rd International Conference on Machine Learning. Springer. pp. 753--760.
- Breiman L (1999). "Prediction Games and Arcing Algorithms". Neural Computation. 11: 1493-1517.
- Mease D, Wyner A (2008). "Evidence Contrary to the Statistical View of Boosting". The Journal of Machine Learning Research. 9: 9-131.
- Buhlmann P, Hothorn T (2007). "Rejoinder: Boosting Algorithms: Regularization, Prediction and Model Fitting". Statistical Science. 22: 516-522.
- Buhlmann P, Yu B (2008). "Response to Mease and Wyner, Evidence Contrary to the Statistical View of Boosting". Journal of Machine Learning Research. 9: 187-194.
- Hastie T, Tibshirani R, Friedman J. (1990). Generalized Additive Models. London: Chapman & Hall.CS1 maint: multiple names: authors list (link)
- 33.0 33.1 33.2 33.3 Friedman JH (2001). "Greedy Function Approximation: A Gradient Boosting Machine". The Annals of Statistics 29: 1189-1232.
- Buhlmann P, Yu B (2003). "Boosting with the L_2 Loss: Regression and Classification". Journal of the American Statistical Association 98: 324--338.
- Buhlmann P (2006). "Boosting for High-Dimensional Linear Models". The Annals of Statistics. 34: 559--583.
- Buhlmann P, Hothorn T (2007). "Boosting Algorithms: Regularization, Prediction and Model Fitting (with Discussion)". Statistical Science. 22: 477-522.
- Schmid M, Hothorn T (2008). "Boosting Additive Models Using Component-Wise P-splines". Computational Statistics Data Analysis. 53: 298--311.
- Hofner B, Mayr A, Robinzonov N, Schmid M (2014). "Model-Based Boosting in R: A Hands-on Tutorial Using the R Package mboost". Computational Statistics. 29: 3--35.
- Tibshirani R (1996). "Regression Shrinkage and Selection via the Lasso". Journal of the Royal Statistical Society - Series B. 58(1): 267--288.
- 40.0 40.1 Mayr A, Hofner B, Schmid M (2012). "The Importance of Knowing When to Stop -- A Sequential Stopping Rule for Component-Wise Gradient Boosting". Methods of Information in Medicine. 51(2): 178-186.
- Hansen MH, Yu B (2001). "Model Selection and the Principle of Minimum Description Length". Journal of the American Statistical Association. 96(454): 746--774.
- Hastie T (2007). "Comment: Boosting Algorithms: Regularization, Prediction and Model Fitting". Statistical Science. 22(4): 513--515.
- Briau, Gérard; Scornet, Erwan (2015). "A Random Forest Guided Tour". arXiv:1511.05741 [math.ST].
- 44.0 44.1 44.2 44.3 44.4 44.5 Breiman, L, (2001). "Random forests.". Machine Learning 45: 5-32.
- "Shape quantization and recognition with randomized trees." (1997). Neural Computation 9: 1545-1588.
- "The random subspace method for constructing decision forests." (1998). Pattern Analysis and Machine Intelligence 20: 832-844.
- Dietterich, T.G, (2000). Multiple Classifier Systems. Springer. pp. 1–15.CS1 maint: extra punctuation (link)
- "On the asymptotics of random forests." (2015a). Journal of Multivariate Analysis, in press.
- 49.0 49.1 Breiman; Friedman; Stone, Olshen (1984). Classification and Regression Trees. Boca Raton: Chapman & Hall/CRC. pp. 1–15.
- 50.0 50.1 50.2 50.3 50.4 "Gene selection and classification of microarray data using random forest." (2006). BMC Bioinformatics 7: 1-13.
- Devroye, L.; Györfi, L.; Lugosi, G. (1996). A Probabilistic Theory of Pattern Recognition. New York: Springer. pp. 1–15.
- "Classification and regression by randomForest." (2002). R News 2: 18-22.
- "Probability machines: consistent probability estimation using nonparametric learning machines." (2012). Methods of Information in Medicine 51: 74-81.
- "Probability estimation with machine learning methods for dichotomous and multicategory outcome: Theory." (2014a). Biometrical Journal 56: 534-563.
- "Probability estimation with machine learning methods for dichotomous and multicategory outcome: Theory." (2014b). Biometrical Journal 56: 564-583.
- 56.0 56.1 Heutte, Bernard.; Adam, S. (2008). "Forest-RK: A new random forest induction method.". In D.-S. Huang, D.C. Wunsch II, D.S. Levine, and K.-H. Jo (ed.). Advanced Intelligent Computing Theories and Applications. With Aspects of Artificial Intelligence. Berlin: Springer. pp. 430–437.CS1 maint: multiple names: editors list (link)
- 57.0 57.1 57.2 "Variable selection using random forests." (2010). Pattern Recognition Letters. 31: 2225-2236.
- 58.0 58.1 "On safari to random jungle: A fast implementation of random forests for high-dimensional data." (2010). Bioinformatics 26: 1752-1758.
- Latinne, P,; Debeir, O.; Decaestecker, C. (2001). "Limiting the number of trees in random forests.". In J.Kittler and F.Roli (ed.). Multiple Classifier Systems. Springer. pp. 178–187.CS1 maint: extra punctuation (link)
- 60.0 60.1 "Consumer credit risk: Individual probability estimates using machine learning." (2013). Expert Systems with Applications 40: 5125-5131.
- Breiman, L. (2003). "Setting up, using, and understanding random forests V3.1" (PDF). Open Publishing. Retrieved April 18, 2024.
Further Reading
- Breiman, Leo (1996). "Bagging predictors". Machine Learning 24 (2): 123–140. doi:.
- Alfaro, E., Gámez, M. and García, N. (2012). "adabag: An R package for classification with AdaBoost.M1, AdaBoost-SAMME and Bagging".
- Kotsiantis, Sotiris (2014). "Bagging and boosting variants for handling classifications problems: a survey". Knowledge Eng. Review 29 (1): 78–100. doi:.
- Boehmke, Bradley; Greenwell, Brandon (2019). "Bagging". Hands-On Machine Learning with R. Chapman & Hall. pp. 191–202. ISBN 978-1-138-49568-5.
Wikipedia References
- Wikipedia contributors. "Bootstrap aggregating". Wikipedia. Wikipedia. Retrieved 17 August 2022.
- Wikipedia contributors. "Boosting (machine learning)". Wikipedia. Wikipedia. Retrieved 17 August 2022.
- Wikipedia contributors. "Random forest". Wikipedia. Wikipedia. Retrieved 17 August 2022.